publications
publications by categories in reversed chronological order.
2024
-
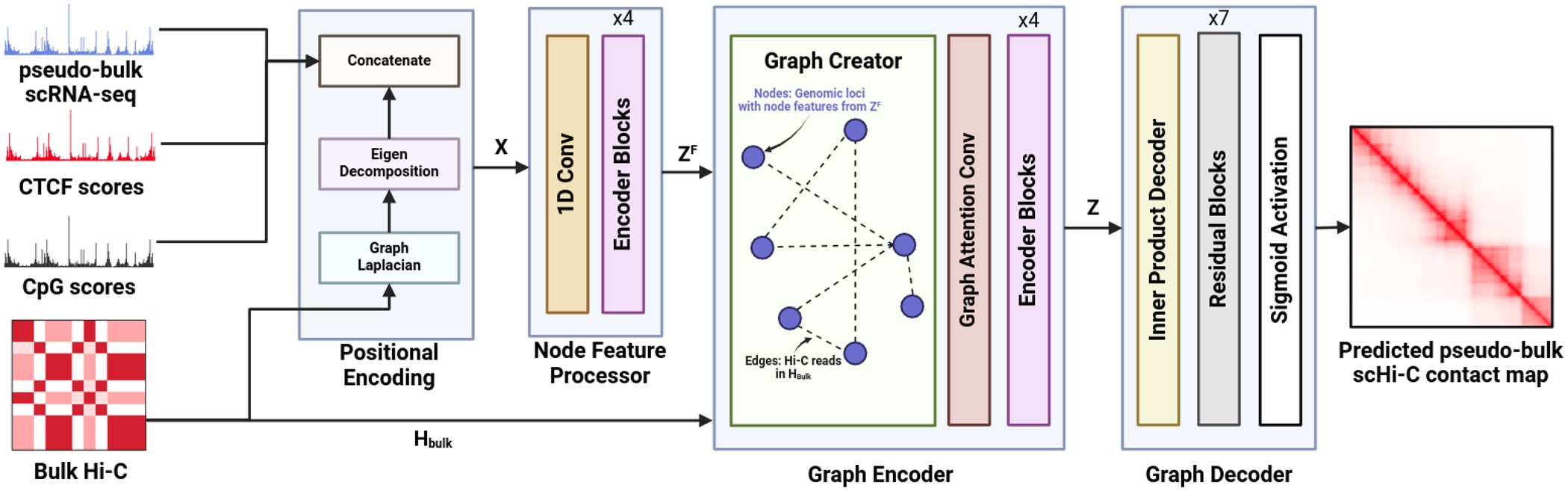 scGrapHiC: Deep Learning-based graph deconvolution for hi-C using single cell gene expressionGhulam Murtaza, Byron Butaney, Justin Wagner, and Ritambhara SinghBioinformatics, Jun 2024
scGrapHiC: Deep Learning-based graph deconvolution for hi-C using single cell gene expressionGhulam Murtaza, Byron Butaney, Justin Wagner, and Ritambhara SinghBioinformatics, Jun 2024Single-cell Hi-C (scHi-C) protocol helps identify cell-type-specific chromatin interactions and sheds light on cell differentiation and disease progression. Despite providing crucial insights, scHi-C data is often underutilized due to the high cost and the complexity of the experimental protocol. We present a deep learning framework, scGrapHiC, that predicts pseudo-bulk scHi-C contact maps using pseudo-bulk scRNA-seq data. Specifically, scGrapHiC performs graph deconvolution to extract genome-wide single-cell interactions from a bulk Hi-C contact map using scRNA-seq as a guiding signal. Our evaluations show that scGrapHiC, trained on seven cell-type co-assay datasets, outperforms typical sequence encoder approaches. For example, scGrapHiC achieves a substantial improvement of in recovering cell-type-specific Topologically Associating Domains over the baselines. It also generalizes to unseen embryo and brain tissue samples. scGrapHiC is a novel method to generate cell-type-specific scHi-C contact maps using widely available genomic signals that enables the study of cell-type-specific chromatin interactions.
-
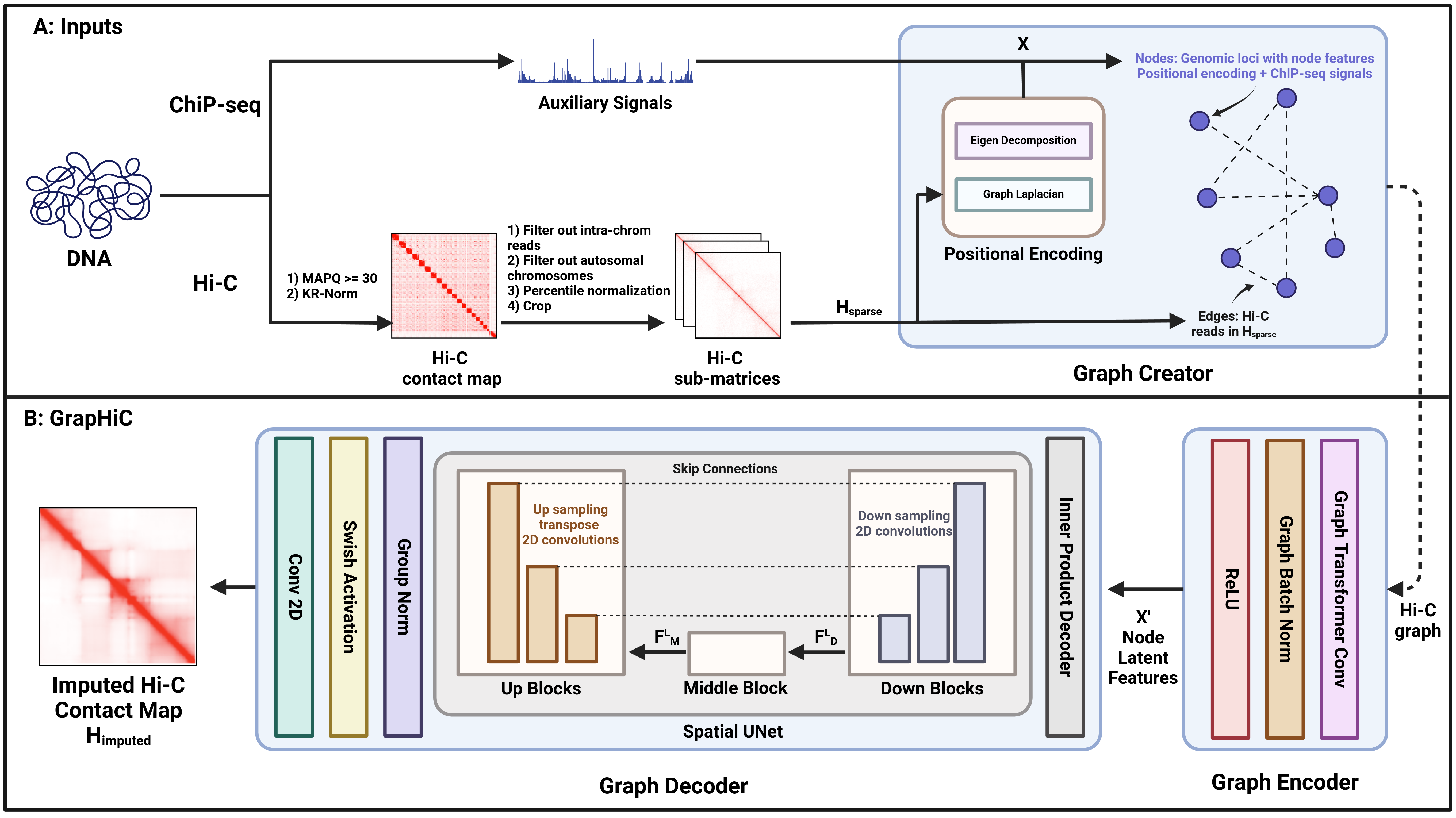 GrapHiC: An integrative graph based approach for imputing missing hi-C readsGhulam Murtaza, Justin Wagner, Justin M. Zook, and Ritambhara SinghbioRxiv, Jul 2024
GrapHiC: An integrative graph based approach for imputing missing hi-C readsGhulam Murtaza, Justin Wagner, Justin M. Zook, and Ritambhara SinghbioRxiv, Jul 2024Hi-C experiments allow researchers to study and understand the 3D genome organization and its regulatory function. Unfortunately, sequencing costs and technical constraints severely restrict access to high-quality Hi-C data for many cell types. Existing frameworks rely on a sparse Hi-C dataset or cheaper-to-acquire ChIP-seq data to predict Hi-C contact maps with high read coverage. However, these methods fail to generalize to sparse or cross-cell-type inputs because they do not account for the contributions of epigenomic features or the impact of the structural neighborhood in predicting Hi-C reads. We propose GrapHiC, which combines Hi-C and ChIP-seq in a graph representation, allowing more accurate embedding of structural and epigenomic features. Each node represents a binned genomic region, and we assign edge weights using the observed Hi-C reads. Additionally, we embed ChIP-seq and relative positional information as node attributes, allowing our representation to capture structural neighborhoods and the contributions of proteins and their modifications for predicting Hi-C reads. Our evaluations show that GrapHiC generalizes better than the current state-of-the-art on cross-cell-type settings and sparse Hi-C inputs. Moreover, we can utilize our framework to impute Hi-C reads even when no Hi-C contact map is available, thus making high-quality Hi-C data more accessible for many cell types.
2023
-
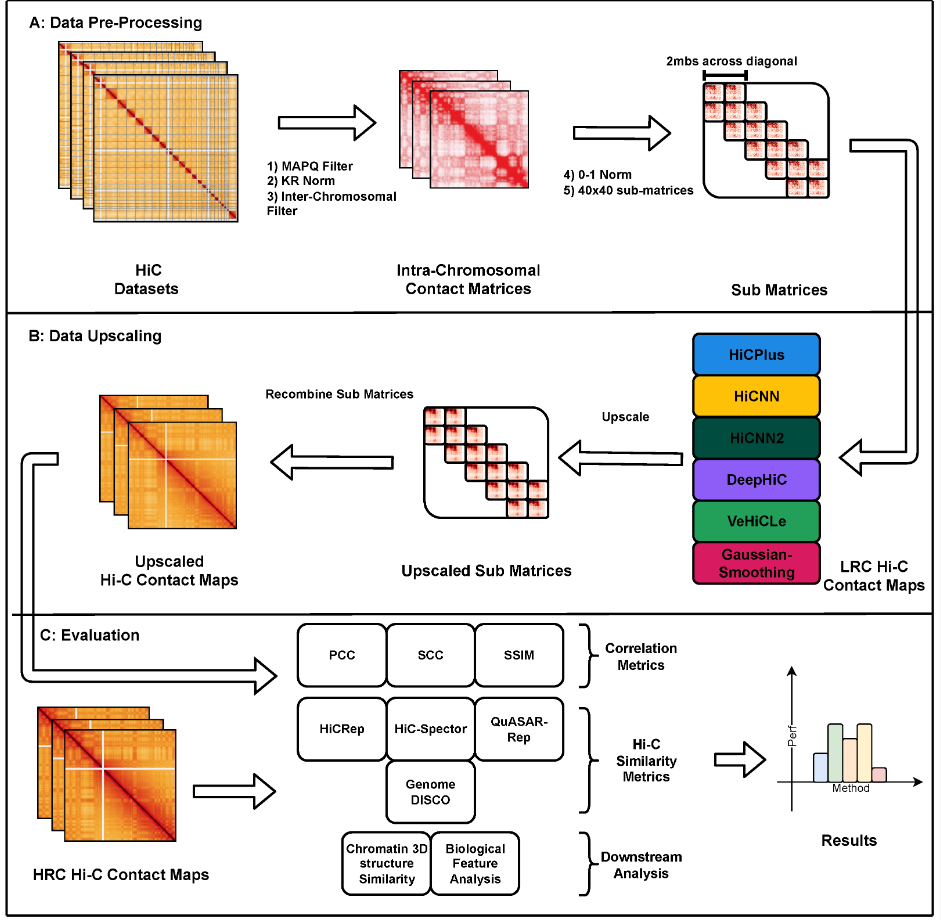 A comprehensive evaluation of generalizability of Deep Learning-based hi-C resolution improvement methodsGhulam Murtaza, Atishay Jain, Madeline Hughes, Justin Wagner, and Ritambhara SinghGenes, Dec 2023
A comprehensive evaluation of generalizability of Deep Learning-based hi-C resolution improvement methodsGhulam Murtaza, Atishay Jain, Madeline Hughes, Justin Wagner, and Ritambhara SinghGenes, Dec 2023Hi-C is a widely used technique to study the 3D organization of the genome. Due to its high sequencing cost, most of the generated datasets are of a coarse resolution, which makes it impractical to study finer chromatin features such as Topologically Associating Domains (TADs) and chromatin loops. Multiple deep learning-based methods have recently been proposed to increase the resolution of these datasets by imputing Hi-C reads (typically called upscaling). However, the existing works evaluate these methods on either synthetically downsampled datasets, or a small subset of experimentally generated sparse Hi-C datasets, making it hard to establish their generalizability in the real-world use case. We present our framework—Hi-CY—that compares existing Hi-C resolution upscaling methods on seven experimentally generated low-resolution Hi-C datasets belonging to various levels of read sparsities originating from three cell lines on a comprehensive set of evaluation metrics. Hi-CY also includes four downstream analysis tasks, such as TAD and chromatin loops recall, to provide a thorough report on the generalizability of these methods. We observe that existing deep learning methods fail to generalize to experimentally generated sparse Hi-C datasets, showing a performance reduction of up to 57%. As a potential solution, we find that retraining deep learning-based methods with experimentally generated Hi-C datasets improves performance by up to 31%. More importantly, Hi-CY shows that even with retraining, the existing deep learning-based methods struggle to recover biological features such as chromatin loops and TADs when provided with sparse Hi-C datasets. Our study, through the Hi-CY framework, highlights the need for rigorous evaluation in the future. We identify specific avenues for improvements in the current deep learning-based Hi-C upscaling methods, including but not limited to using experimentally generated datasets for training.
2021
-
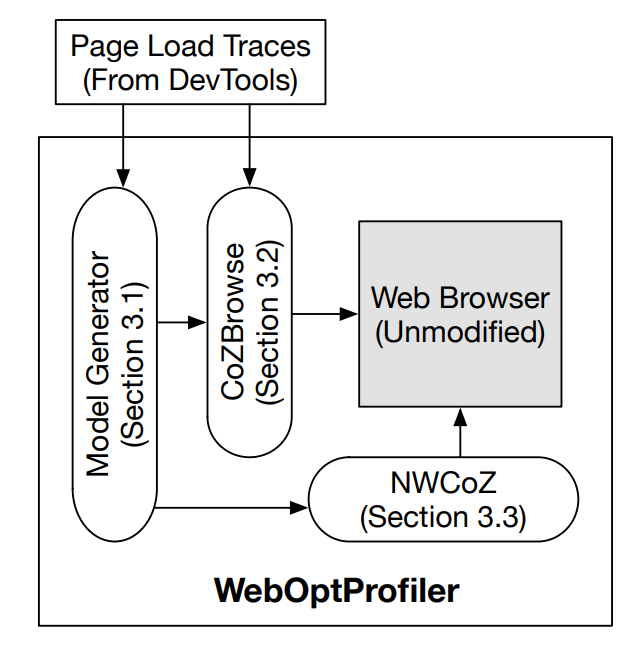 WebOptProfiler: Providing performance clarity for Mobile Webpage OptimizationsGhulam Murtaza, and Theophilus A. BensonProceedings of the 22nd International Workshop on Mobile Computing Systems and Applications, Feb 2021
WebOptProfiler: Providing performance clarity for Mobile Webpage OptimizationsGhulam Murtaza, and Theophilus A. BensonProceedings of the 22nd International Workshop on Mobile Computing Systems and Applications, Feb 2021Despite decades of research on mobile webpage optimizations, little is known about how these optimizations interoperate. Moreover, there has been little systematic work to understand the scenarios wherein combinations of these optimizations excel. Without a comprehensive understanding of how these optimizations compose with each other and under what conditions they excel, operators cannot determine which optimizations to adopt, and, similarly, developers do not know where to focus their efforts. In this paper, we argue that developers should be required to evaluate and characterize the broader interactions between their proposed optimizations and other optimizations - this is in addition to demonstrating the potential benefits of their approach. To aide developers in characterizing these broader interactions, we propose an analytical model which decomposes web optimizations into virtual speedup functions that operate on well-understood browser processing phases (e.g., processing, rendering, layout, etc., for an object) and we present a web browser-oriented causal profiler which empirically explores interactions between optimizations by using their analytical models to speed up different parts of the Browser during a page load. Our system, WebOptProfiler, identifies and addresses practical issues in extending causal profiling to the webpage optimization domain and provides an algorithm for extracting an analytical model from readily available browser traces.
2020
-
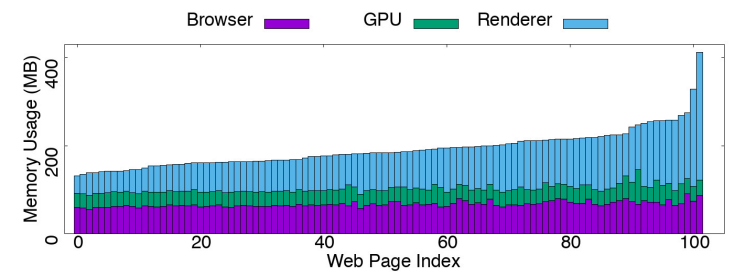 Mobile web browsing under memory pressureIhsan Ayyub Qazi, Zafar Ayyub Qazi, Theophilus A. Benson, Ghulam Murtaza, Ehsan Latif, Abdul Manan, and Abrar TariqACM SIGCOMM Computer Communication Review, Oct 2020
Mobile web browsing under memory pressureIhsan Ayyub Qazi, Zafar Ayyub Qazi, Theophilus A. Benson, Ghulam Murtaza, Ehsan Latif, Abdul Manan, and Abrar TariqACM SIGCOMM Computer Communication Review, Oct 2020Mobile devices have become the primary mode of Internet access. Yet, differences in mobile hardware resources, such as device memory, coupled with the rising complexity of Web pages can lead to widely different quality of experience for users. In this work, we analyze how device memory usage affects Web browsing performance. We quantify the memory footprint of popular Web pages over different mobile devices, mobile browsers, and Android versions, analyze the induced memory distribution across different browser components (e.g., JavaScript engine and compositor), investigate how performance gets impacted under memory pressure and propose optimizations to reduce the memory footprint of Web browsing. We show that these optimizations can improve performance and reduce chances of browser crashes in low memory scenarios.
-
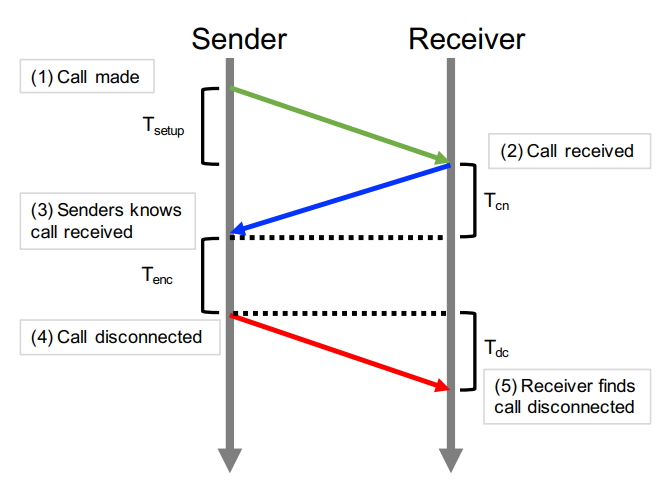 Missit: Using missed calls for free, extremely low bit-rate communication in developing regionsFahad R. Dogar, Ihsan Ayyub Qazi, Ali Raza Tariq, Ghulam Murtaza, Abeer Ahmad, and Nathan StockingProceedings of the 2020 CHI Conference on Human Factors in Computing Systems, Apr 2020
Missit: Using missed calls for free, extremely low bit-rate communication in developing regionsFahad R. Dogar, Ihsan Ayyub Qazi, Ali Raza Tariq, Ghulam Murtaza, Abeer Ahmad, and Nathan StockingProceedings of the 2020 CHI Conference on Human Factors in Computing Systems, Apr 2020Mobile devices have become the primary mode for Internet access in developing countries. Yet typical data plans and SMS costs can be overwhelming for low income users in these countries. In this paper, we explore the design and usability of a free but extremely low bit rate communication channel to address this challenge. We propose MissIt, a data communication channel that uses missed calls to transmit messages between phones, thereby sacrificing performance in exchange for low cost. While the data rate of MissIt is extremely low (<1bps), our prototype implementation and small scale user studies explore the feasibility of this idea for different types of messaging scenarios. Our results show that MissIt could be a viable option for messaging scenarios that require short, pre-determined responses (e.g., survey questions) while for traditional SMS-style messaging, a suitable user interface and other customizations are likely required to make it a viable option for users.