GrapHiC
Predicting high-quality Hi-C contact maps from sparse contact maps
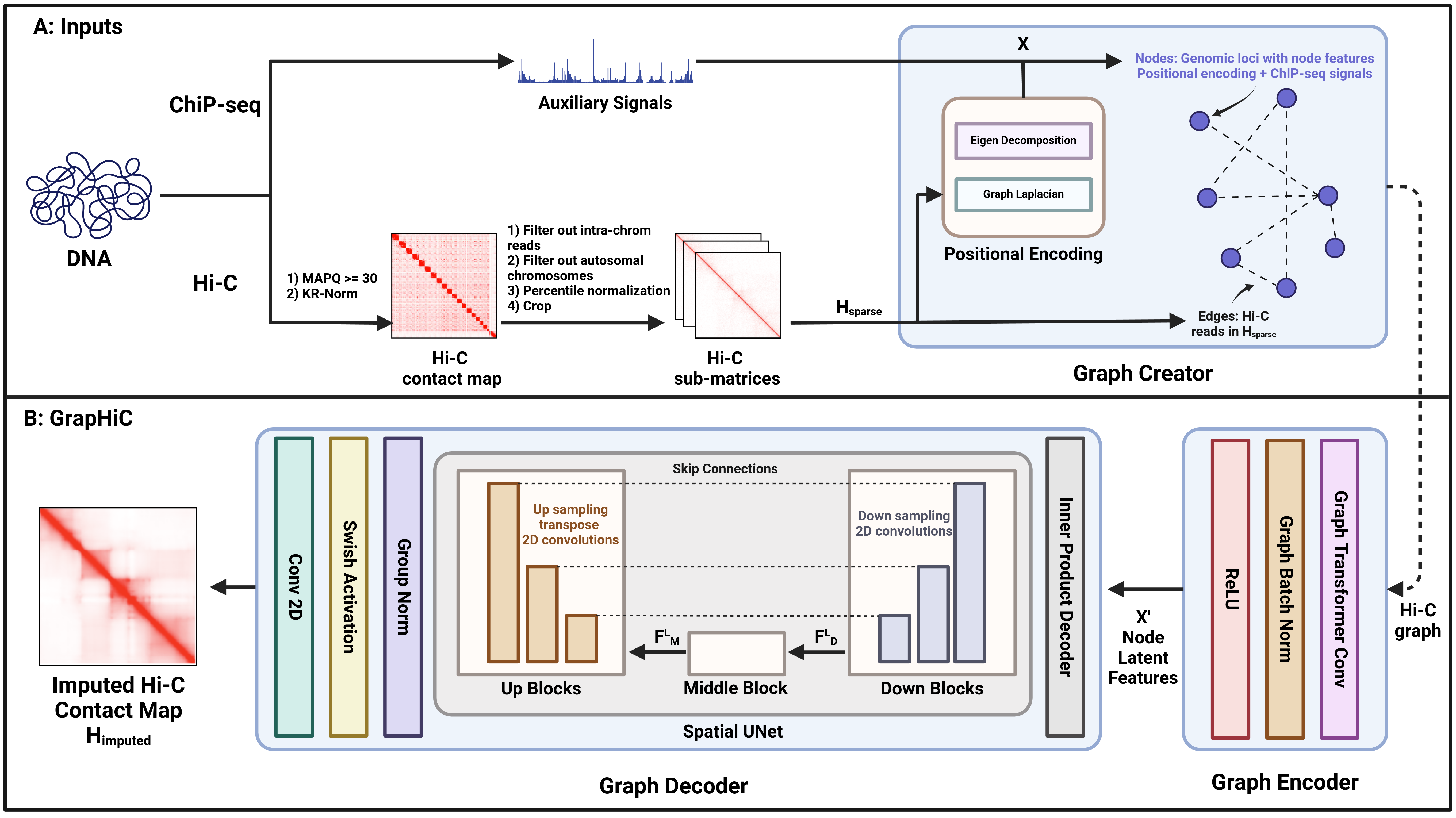
Hi-C experiments allow researchers to study and understand the 3D genome organization and its regulatory function. Unfortunately, sequencing costs and technical constraints severely restrict access to high-quality Hi-C data for many cell types. Existing frameworks rely on a sparse Hi-C dataset or cheaper-to-acquire ChIP-seq data to predict Hi-C contact maps with high read coverage. However, these methods fail to generalize to sparse or cross-cell-type inputs because they do not account for the contributions of epigenomic features or the impact of the structural neighborhood in predicting Hi-C reads. We propose GrapHiC, which combines Hi-C and ChIP-seq in a graph representation, allowing more accurate embedding of structural and epigenomic features. Each node represents a binned genomic region, and we assign edge weights using the observed Hi-C reads. Additionally, we embed ChIP-seq and relative positional information as node attributes, allowing our representation to capture structural neighborhoods and the contributions of proteins and their modifications for predicting Hi-C reads. We show that GrapHiC generalizes better than the current state-of-the-art on cross-cell-type settings and sparse Hi-C inputs. Moreover, we can utilize our framework to impute Hi-C reads even when no Hi-C contact map is available, thus making high-quality Hi-C data accessible for many cell types.
The code is available at: https://github.com/rsinghlab/GrapHiC