scGrapHiC
Predicting pseudo-bulk scHi-C from scRNA-seq
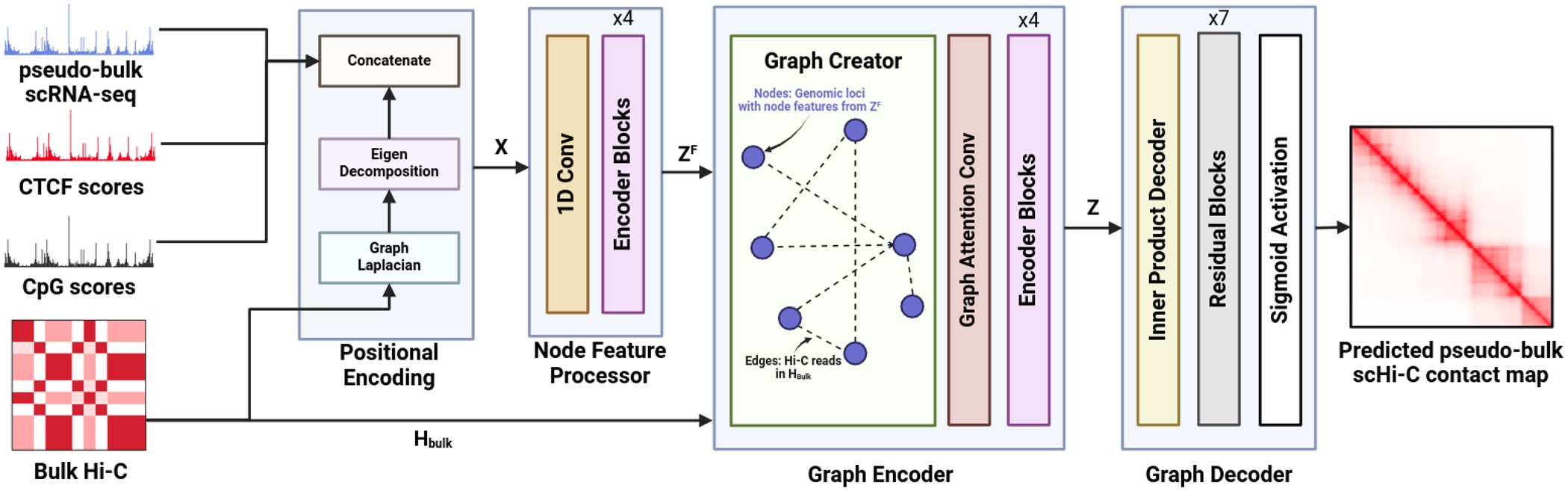
Single-cell Hi-C (scHi-C) protocol helps identify cell-type-specific chromatin interactions and sheds light on cell differentiation and disease progression. Despite providing crucial insights, scHi-C data is often underutilized due to the high cost and the complexity of the experimental protocol. We present a deep learning framework, scGrapHiC, that predicts pseudo-bulk scHi-C contact maps using pseudo-bulk scRNA-seq data. Specifically, scGrapHiC performs graph deconvolution to extract genome-wide single-cell interactions from a bulk Hi-C contact map using scRNA-seq as a guiding signal. Our evaluations show that scGrapHiC, trained on 7 cell-type co-assay datasets, outperforms typical sequence encoder approaches. For example, scGrapHiC achieves a substantial improvement of $23.2\%$ in recovering cell-type-specific Topologically Associating Domains over the baselines. It also generalizes to unseen embryo and brain tissue samples. scGrapHiC is a novel method to generate cell-type-specific scHi-C contact maps using widely available genomic signals that enables the study of cell-type-specific chromatin interactions.
The code is available at: https://github.com/rsinghlab/scGrapHiC The full manuscript is available at: https://academic.oup.com/bioinformatics/article/40/Supplement_1/i490/7700876